load required packages
library(raster)
library(rasterVis)
library(rgee)
library(sf)
library(tidyverse)
Getting Landcover data from Google Earth Engine
library(rgee)
ee_Initialize(drive = T)
roi <- ee$Geometry$Rectangle(76.8100, 30.6676, 77.0010, 30.8084)
landcover2020 <- ee$ImageCollection("ESA/WorldCover/v100")$
first()$
clip(roi)
task_img <- ee_image_to_drive(
image = landcover2020,
folder = "rgee_backup",
fileFormat = "GEO_TIFF",
region = roi,
fileNamePrefix = "landcover_2020"
)
task_img$start()
ee_monitoring(task_img)
img <- ee_drive_to_local(task = task_img, dsn = "data/lc2020.tif")
googledrive::drive_deauth()
rm(list = ls())
base R
mwls_lc <- raster("data/lc2020.tif")
plot(mwls_lc)


lattice levelplot
library(raster)
library(rasterVis)
mwls_lc <- raster("data/lc2020.tif")
levels(mwls_lc) <- data.frame(
ID = c(seq(10, 60, 10), 80),
landcover = c("Trees", "Shrubland", "Grassland", "Cropland", "Built-up",
"Barren/Sparse \n Vegetation", "Open water")
)
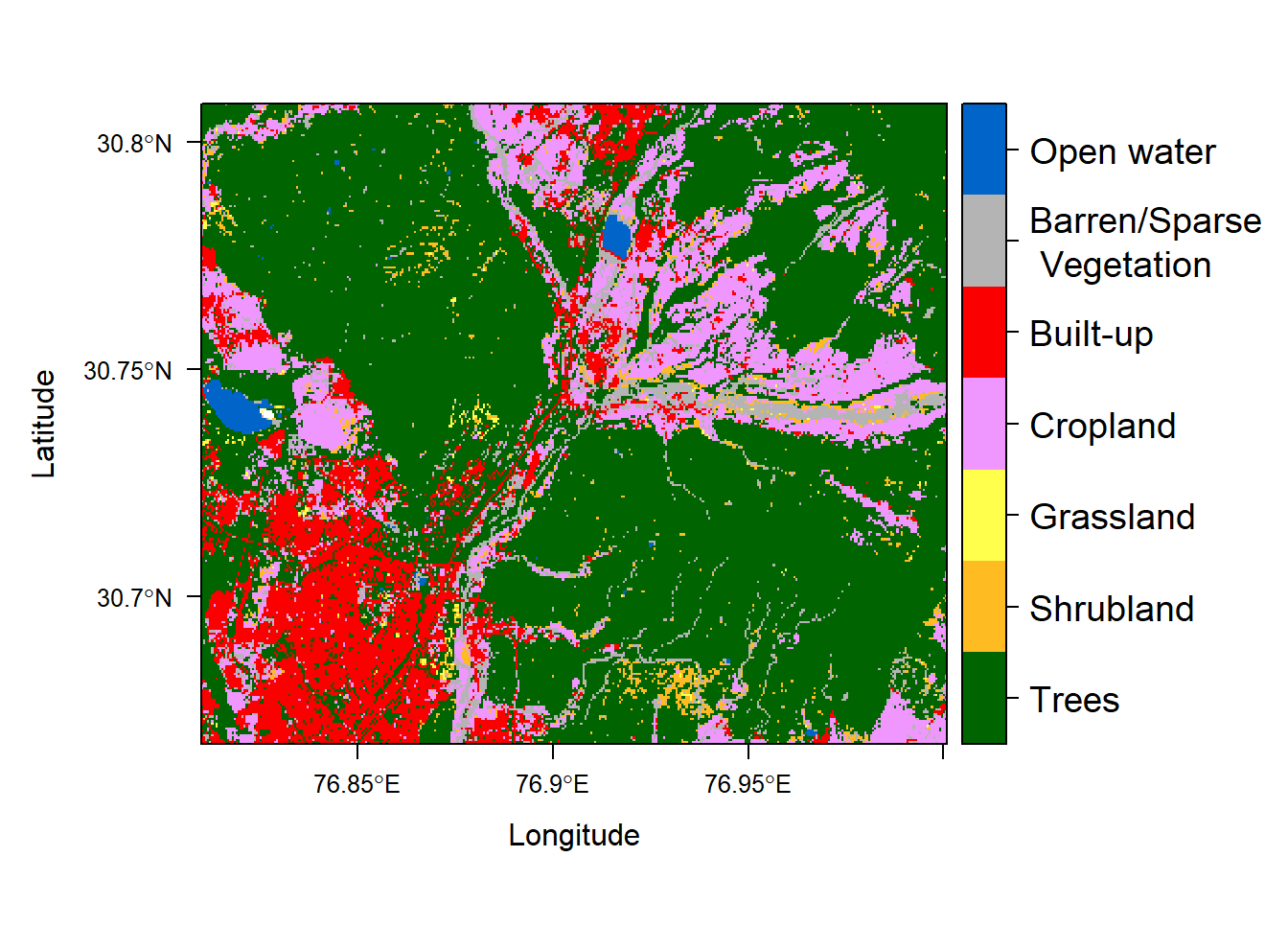
levelplot(mwls_lc, att='landcover',
col.regions = c("#006400", "#FFBB22", "#FFFF4C", "#F096FF", "#FA0000",
"#B4B4B4", "#0064C8"),
colorkey = list(height = 1, labels = list(cex = 1.2)))
ggplot
mwls_lc <- raster("data/lc2020.tif")
rasterdf <- function(x, aggregate = 1){
resampleFactor <- aggregate
inputRaster <- x
inCols <- ncol(inputRaster)
inRows <- nrow(inputRaster)
resampledRaster <- raster(ncol = (inCols / resampleFactor),
nrow = (inRows / resampleFactor))
extent(resampledRaster) <- extent(inputRaster)
y <- resample(inputRaster, resampledRaster, method='ngb')
coords <- xyFromCell(y, seq_len(ncell(y)))
dat <- stack(as.data.frame(getValues(y)))
names(dat) <- c('value', 'variable')
dat <- cbind(coords, dat)
dat
}
mwls_df <- rasterdf(mwls_lc)
LCcodes <- c(seq(10, 90, 10), 95, 100)
LCnames <- c("Trees", "Shrubland", "Grassland", "Cropland", "Built-up",
"Barren/Sparse Vegetation", "Snow and Ice", "Open water",
"Herbaceous wetland", "Mangroves", "Moss and lichen")
LCcolors <- attr(mwls_lc, "legend")@colortable[LCcodes + 1]
names(LCcolors) <- as.character(LCcodes)
ggplot(data = mwls_df) +
geom_raster(aes(x = x, y = y, fill = as.character(value))) +
scale_fill_manual(name = "Land cover",
values = LCcolors[-c(7, 9:11)],
labels = LCnames[-c(7, 9:11)],
na.translate = FALSE) +
coord_sf(expand = F) +
theme(axis.title.x = element_blank(),
axis.title.y = element_blank(),
panel.background = element_rect(fill = "white", color = "black"))

rm(list = ls())
